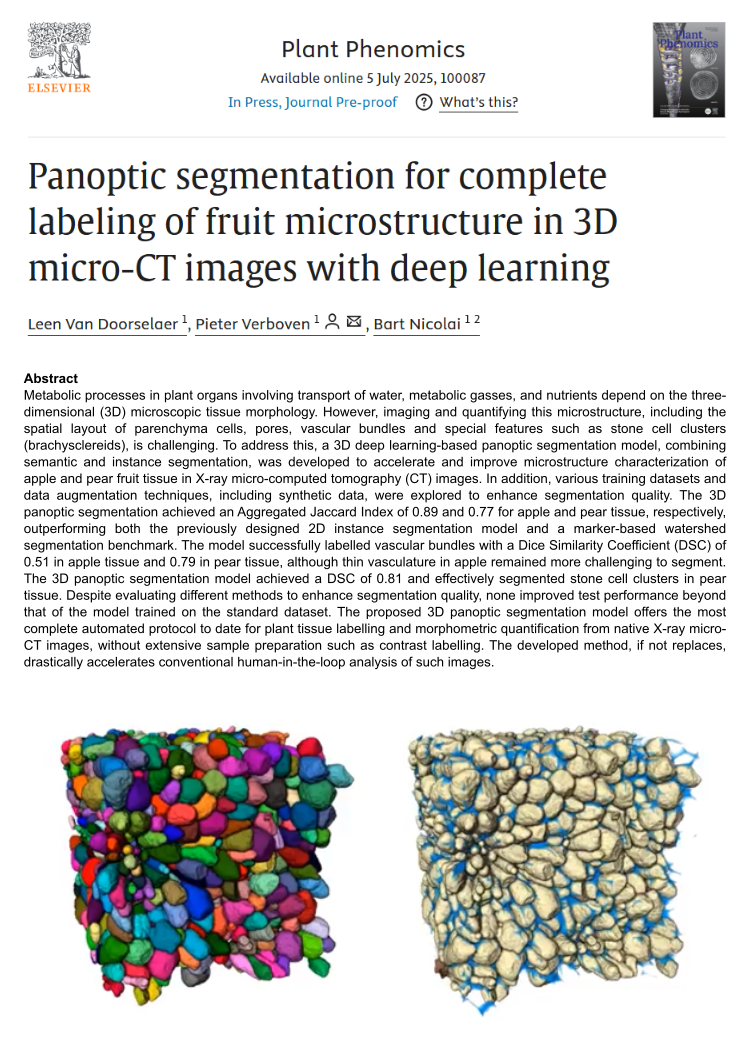
Metabolic processes in plant organs involving transport of water, metabolic gasses, and nutrients depend on the three-dimensional (3D) microscopic tissue morphology. However, imaging and quantifying this microstructure, including the spatial layout of parenchyma cells, pores, vascular bundles and special features such as stone cell clusters (brachysclereids), is challenging. To address this, a 3D deep learning-based panoptic segmentation model, combining semantic and instance segmentation, was developed to accelerate and improve microstructure characterization of apple and pear fruit tissue in X-ray micro-computed tomography (CT) images. In addition, various training datasets and data augmentation techniques, including synthetic data, were explored to enhance segmentation quality. The 3D panoptic segmentation achieved an Aggregated Jaccard Index of 0.89 and 0.77 for apple and pear tissue, respectively, outperforming both the previously designed 2D instance segmentation model and a marker-based watershed segmentation benchmark. The model successfully labelled vascular bundles with a Dice Similarity Coefficient (DSC) of 0.51 in apple tissue and 0.79 in pear tissue, although thin vasculature in apple remained more challenging to segment. The 3D panoptic segmentation model achieved a DSC of 0.81 and effectively segmented stone cell clusters in pear tissue. Despite evaluating different methods to enhance segmentation quality, none improved test performance beyond that of the model trained on the standard dataset. The proposed 3D panoptic segmentation model offers the most complete automated protocol to date for plant tissue labelling and morphometric quantification from native X-ray micro-CT images, without extensive sample preparation such as contrast labelling. The developed method, if not replaces, drastically accelerates conventional human-in-the-loop analysis of such images.